Problem Statement¶
An education company named X Education sells online courses to industry professionals. The company markets its courses on several websites and search engines like Google. Once these people land on the website, they might browse the courses or fill up a form for the course or watch some videos. When these people fill up a form providing their email address or phone number, they are classified to be a lead.
Once these leads are acquired, employees from the sales team start making calls, writing emails, etc. Through this process, some of the leads get converted while most do not. The typical lead conversion rate at X education is around 30%.
To make this process more efficient, the company wishes to identify the most potential leads, also known as ‘Hot Leads’. If they successfully identify this set of leads, the lead conversion rate should go up as the sales team will now be focusing more on communicating with the potential leads rather than making calls to everyone. A typical lead conversion process can be represented using the following funnel:
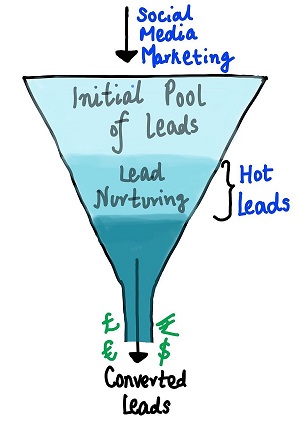
"The company requires a model that will assign a lead score to each of the leads such that the customers with higher lead score have a higher conversion chance and the customers with lower lead score have a lower conversion chance."
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from matplotlib import style
import seaborn as sns
style.use('ggplot')
import statsmodels.api as sm
from statsmodels.stats.outliers_influence import variance_inflation_factor
from sklearn.preprocessing import MinMaxScaler
from sklearn.decomposition import PCA
from sklearn.feature_selection import RFE
from sklearn.model_selection import train_test_split, GridSearchCV, cross_validate
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import confusion_matrix, roc_curve, roc_auc_score, precision_score, recall_score, precision_recall_curve, f1_score, accuracy_score
import warnings
warnings.filterwarnings("ignore")
pd.set_option("display.max_columns", 50)
# Importing data
df = pd.read_csv("D:\\DataSets\\UPGRAD\\Assignments\\Lead+Scoring+Case+Study\\Leads.csv")
df.head()
df.info()
df.describe()
Data Cleaning¶
# Standardising column names
df.columns = ['_'.join(name.lower().split()[:3]) for name in df.columns]
#standardising categorical values
cat_cols = df.select_dtypes(include=['object']).columns
df[cat_cols] = df[cat_cols].apply(lambda x: x.str.lower().str.replace(' ', '_').str.strip())
# Taking a look at all unique values for every column to check for abnormalities/inconsistencies
for col in df.columns[2:]:
print(col.upper(), end=': ')
print(df[col].unique())
print()
Issues by column:¶
By going through the above unique entires of every column, we have observed the following inconsistencies:
lead_source -> WeLearn and WeLearnblog_home are one and the same. Facebook and social media are two different categories.
country -> "Australia", the asian Countries and "Asia/Pacific Region" are two different categories.
tags -> "invalid number" and "wrong number given" are one and the same.
last_activity and last_notable_activity are highly correlated. One of them should be dropped to avoid multi-collinearity.
Other potential issues:¶
- Many columns exist with only a single category (Redundant columns).
- same category percieved as different due to not matching case. (ex. Google -> google)
- The value
Selectis equivalent to NaN. - Possible overlaps may exist in City column.
# removing redundant columns
df.drop(['prospect_id', 'lead_number', 'receive_more_updates', 'update_me_on', 'get_updates_on', 'i_agree_to', 'last_activity'], axis=1, inplace=True)
# Replacing all 'select' values with NaN
df = df.replace('select', np.nan)
# replacing "wrong number given" with "invalid number"
df.tags = df.tags.replace("wrong_number_given", "invalid_number")
# Cleaning "lead_source column"
value_counts = df.lead_source.value_counts()
# Values with frequency less than 30 are classified under "others"
df.lead_source = df.lead_source.replace(value_counts[value_counts < 30].index, "others")
# Cleaning "country"
value_counts = df.country.value_counts()
# We will categorise Country in binary as 'India' and 'Other'
df.country = df.country.replace(df.country[df.country != 'india'].dropna().unique(), "others")
Dealing with NaN values¶
# Percentage of NA values in every column
round(df.isna().sum().sort_values(ascending=False)/len(df)*100, 2)
# Column "how_did_you" has too many NA values that can not be imputed. It is better to drop the column.
df.drop('how_did_you', axis=1, inplace=True)
# Column "lead_profile" has too many NA values that can not be imputed.
print(df.lead_profile.value_counts())
print("\n\nWe could convert the column to binary by defining whether it is a 'potential_lead' or not.")
print(f"\nIf made binary variable:\nis_potential_lead: {df.lead_profile.value_counts()[0]}\nis_not_lead: {len(df) - df.lead_profile.value_counts()[0]}\n")
print("But given the high frequency of NA values, There is a lot of uncertainty whether those values are leads or not. This forces us to drop the column altogether.")
df.drop('lead_profile', axis=1, inplace=True)
# Column "lead_quality" has 52% NA values. We'll classify them as "not_sure".
df.lead_quality.fillna("not_sure", inplace=True)
print(df.lead_quality.value_counts())
# Asymmetrique scores
for col in ['profile_score', 'activity_score']:
sns.distplot(df['asymmetrique_'+col].dropna())
plt.show()
print(f"Both scores roughly follow standard distribution, therefore imputation can be done by median value.\nThough instead of using scores, we shall use the categorical index columns derived from these scores.\nMissing values will be filled by most frequently occuring category.")
# Dropping Scores (Only Indexes will be used)
df.drop(['asymmetrique_profile_score', 'asymmetrique_activity_score'], axis=1, inplace=True)
# Removing initial numbers from category naming
df['asymmetrique_profile_index'] = df['asymmetrique_profile_index'].str[3:]
df['asymmetrique_activity_index'] = df['asymmetrique_activity_index'].str[3:]
# Replacing nan with most commonly occuring category
df['asymmetrique_activity_index'].fillna(df['asymmetrique_activity_index'].mode().values[0], inplace=True)
df['asymmetrique_profile_index'].fillna(df['asymmetrique_profile_index'].mode().values[0], inplace=True)
# Imputing for 'Country' and 'City'
print("\nWe'll first try understanding how cities are labelled for different country categories\n")
for country in df.country.dropna().unique():
print(country.upper())
print(df[df.country == country]['city'].value_counts())
print()
print("It looks like customers outside India are wrongly marked as being from domestic cities.\n")
print("\nLets look at how countries are labeled w.r.t city categories\n")
for city in df.city.dropna().unique():
print(city.upper())
print(df[df.city == city]['country'].value_counts())
print()
# We will make a new city label ('international') for countries labeled as 'others'
df.city[df.country == "others"] = df.city[df.country == "others"].replace(df.city[df.country == "others"].unique(), 'international')
# We'll also assume that rows with a domestic city label will have to have country label as 'india'.
df.country[(df.city != "international") & ~df.city.isna()] = 'india'
# We'll assume that rows with a country label 'india 'will have most frequently occuring city label.
df.city[(df.country == "india") & (df.city.isna())] = 'mumbai'
print(f"There are {len(df[(df.city.isna()) & (df.country.isna())])} entries where both country and city are NA")
print("We'll mark these as 'unknown'")
df[['city', 'country']] = df[['city', 'country']].fillna('unknown', axis=1)
# Few columns have less than 2% na values. We can afford to drop their respective rows altogehter.
df.dropna(subset=['totalvisits', 'page_views_per', 'lead_source'], inplace=True)
# There are too many NA values with no logical way to impute in these columns so we will drop them entirely
df.drop(['specialization', 'tags', 'what_matters_most', 'what_is_your'] , axis=1, inplace=True)
round(df.isna().sum().sort_values(ascending=False), 2)
Some features have hierarchial categories, which requires ordinal encoding. But then the features will not be scale invariant. It is thus better to one-hot encode all categorical variables.
# dummy encoding for the categorical variables
dummies = pd.get_dummies(df.select_dtypes(include=['object']), drop_first=True)
# getting the cleaned df
clean_df = df.drop(df.select_dtypes(include=['object']).columns, axis=1)
clean_df = pd.concat([clean_df, dummies], axis=1)
clean_df.head()
print(f"{round(len(clean_df)/9239*100,2)}% data has been retained after data cleaning.")
EDA¶
Correlations¶
# visualizing correlation by heatmap
plt.figure(figsize=(14, 10))
sns.heatmap(clean_df.corr())
plt.show()
# heatmap of only continuous variables
sns.heatmap(clean_df[clean_df.columns[:4]].corr())
plt.show()
# columns pairs in order of highest absolute correlation
clean_df.corr().abs().unstack().sort_values(ascending=False).drop_duplicates().head(12)
# Dropping variables with high multi-collinearity
clean_df.drop(['lead_source_facebook', 'lead_origin_lead_add_form', 'lead_source_olark_chat'], axis=1, inplace=True)
# Top 5 features correlated with target variable
clean_df.corr()['converted'].abs().sort_values(ascending=False).head(6)[1:]
Univariate and Outlier Analysis¶
#Histogram
def plot_bars():
plt.figure(figsize=(12, 4))
plt.subplot(121)
sns.distplot(clean_df['total_time_spent'])
plt.subplot(122)
sns.distplot(clean_df['totalvisits'])
plt.tight_layout()
plt.show()
plot_bars()
# Boxplots before outlier removal
num_df = clean_df[['converted', 'totalvisits', 'page_views_per', 'total_time_spent']]
def plot_boxes():
plt.figure(figsize=(12, 4))
plt.subplot(121)
sns.boxplot(data=clean_df, x='total_time_spent')
plt.subplot(122)
sns.boxplot(data=clean_df, x='totalvisits')
plt.tight_layout()
plt.show()
plt.figure(figsize=(6, 6))
box_long = pd.melt(num_df.drop('total_time_spent', axis=1), id_vars='converted')
sns.boxplot(x='converted', y='value', hue='variable', data=box_long)
plt.show()
plot_boxes()
# Removing outliers
out_df = num_df.drop('converted', axis=1)
q = out_df.quantile(0.97)
out_df = out_df[out_df < q]
out_df = out_df.dropna()
clean_df = clean_df.loc[out_df.index].reset_index(drop=True)
num_df = num_df.loc[out_df.index].reset_index(drop=True)
print(f"{round(len(clean_df)/9239*100,2)}% data has been retained after outlier removal.")
# After Outlier removal
plot_bars()
plot_boxes()
Bivariate Analysis¶
sns.pairplot(data=clean_df, vars=clean_df.columns[1:4], hue='converted', kind='reg', height=3,
plot_kws={'scatter_kws': {'alpha': 0.1}})
plt.show()
Building the Predictive Model¶
Splitting into Train and Test¶
We will perform stratified split to prevent the effect of class imbalance of target variable, thus preventing bias.
# Stratified Train Test Split
X = clean_df.drop('converted', axis=1)
y = clean_df['converted']
X_train, X_test, y_train, y_test = train_test_split(X, y, stratify=y, test_size=0.33, random_state=42)
Scaling (Normalisation)¶
We will perform Min Max scaling on the Continuous numerical variables
# Min Max Scaling
scaler = MinMaxScaler()
cols = X_train.columns
scaled_X_train = pd.DataFrame(scaler.fit_transform(X_train[cols[:3]]), columns=cols[:3])
scaled_X_train = pd.concat([scaled_X_train, X_train.drop(cols[:3], axis=1).reset_index(drop=True)], axis=1)
scaled_X_test = pd.DataFrame(scaler.transform(X_test[cols[:3]]), columns=cols[:3])
scaled_X_test = pd.concat([scaled_X_test, X_test.drop(cols[:3], axis=1).reset_index(drop=True)], axis=1)
PCA¶
# Generating principal components of continuous variables
pca = PCA(random_state=42)
decomp_df = pca.fit_transform(scaled_X_train[cols[:3]])
pc_frame = pd.DataFrame({'Features': cols[:3], 'PC1': pca.components_[0], 'PC2': pca.components_[1], 'PC3': pca.components_[2]})
pc_frame
# Plotting 2 principal compnonents for the numeric variables
plt.figure(figsize=(12, 6))
sns.scatterplot(data=pc_frame, x='PC1', y='PC2')
for i, text in enumerate(pc_frame.Features):
plt.annotate(text, (pc_frame.PC1[i], pc_frame.PC2[i]))
plt.show()
# No correlation exists between the PCs
decomp_df = pd.DataFrame(decomp_df, columns=['PC1', 'PC2', 'PC3'])
sns.heatmap(decomp_df.corr())
plt.show()
# Pairplot Of all 3 PCs
sns.pairplot(data=pd.concat([decomp_df, y_train.reset_index(drop=True)], axis=1),
vars=['PC1', 'PC2', 'PC3'], hue='converted', kind='reg', height=3,
plot_kws={'scatter_kws': {'alpha': 0.1}})
plt.show()
# We'll use all 3 principal components
scaled_X_train = pd.concat([scaled_X_train.reset_index(drop=True), decomp_df], axis=1)
# applying to test set
decomp_test = pca.transform(scaled_X_test[cols[:3]])
decomp_test = pd.DataFrame(decomp_test, columns=['PC1', 'PC2', 'PC3'])
scaled_X_test = pd.concat([scaled_X_test.reset_index(drop=True), decomp_test], axis=1)
Recurssive Feature Elimination and Cross Validation¶
By iterating through number of features we will see which model gives highest result.
def optimal_features(min, max):
opt = list()
for features in range(min, max):
log_reg = LogisticRegression(C=2, random_state=42)
rfe = RFE(log_reg, features)
rfe.fit(scaled_X_train, y_train)
cols = scaled_X_train.columns[rfe.support_]
# Cross Validation
scores = cross_validate(log_reg, scaled_X_train[cols], y_train, return_train_score=True, cv=5, scoring=['accuracy'])
opt.append((features, scores['test_accuracy'].mean()))
opt = np.array(opt)
return opt, opt[opt[:, 1].argmax()]
feat_array, opt_features = optimal_features(10, 50)
Here we'll plot how accuracy changes with number of features considered.
plt.figure(figsize=(12, 6))
plt.plot(feat_array[:, 0], feat_array[:, 1])
plt.xlabel("Number of Features")
plt.ylabel("Cross Validated Mean Accuracy")
plt.show()
print(f"Optimal number of features to use is {opt_features[0]} which gives {opt_features[1]} accuracy.")
# RFE
log_reg = LogisticRegression(C=2, random_state=42)
rfe = RFE(log_reg, int(opt_features[0]))
rfe.fit(scaled_X_train, y_train)
cols = scaled_X_train.columns[rfe.support_]
print(f"The columns we'll be using are:\n\n{cols}")
VIF¶
# Manual Elimination by VIF analysis
def get_vif():
vif = pd.DataFrame()
vif['Features'] = scaled_X_train[cols].columns
vif['VIF'] = [variance_inflation_factor(scaled_X_train[cols].values, i) for i in range(scaled_X_train[cols].shape[1])]
vif['VIF'] = round(vif['VIF'], 2)
vif = vif.sort_values(by = "VIF", ascending = False)
return vif
get_vif()
The VIF of total_time_spent is too high. We will eliminate the feature and check the VIF again.
cols = cols.drop('total_time_spent')
get_vif().head()
The factors for the columns are now in check, we can proceed further with our analysis.
Assesing the model with statsmodel¶
to_drop = ['last_notable_activity_resubscribed_to_emails', 'lead_source_welingak_website']
X_train_sm = sm.add_constant(scaled_X_train[cols].drop(to_drop, axis=1))
logm2 = sm.GLM(list(y_train), X_train_sm, family=sm.families.Binomial())
res = logm2.fit()
res.summary()
Fitting the model¶
# Cross Validation
scores = cross_validate(log_reg, scaled_X_train[cols], y_train, return_train_score=True, cv=5, scoring=['accuracy'])
print(f"Cross validated mean accuracy: {round(scores['test_accuracy'].mean(), 3)}")
log_reg.fit(scaled_X_train[cols], y_train)
pred = log_reg.predict(scaled_X_train[cols])
prob_est = log_reg.predict_proba(scaled_X_train[cols])
Measuring Model Performance¶
The data available at hand has class imbalance and therefore accuracy is not a good enough metric to measure if model is good enough.
Sensitivity (Recall) tells us what percentage of leads that were converted, were correctly identified as converted.
Specificity tells is what percentage of leads that were NOT converted were correctly identified.
Precision is, given a positive test result, the sample is positive.
If correctly identifying positives is important for us, then we should choose a model with higher Sensitivity. However, if correctly identifying negatives is more important, then we should choose specificity as the measurement metric.
F1 score is the weighted average of the precision and recall, and is a good metric to hold the model against.
def draw_roc(actual_values, probability_estimates):
fpr, tpr, thresholds = roc_curve(actual_values, probability_estimates, drop_intermediate=False)
auc_score = roc_auc_score(actual_values, probability_estimates)
plt.figure(figsize=(5, 5))
plt.plot(fpr, tpr, label=f"ROC curve (area = {round(auc_score, 2)})")
plt.plot([0, 1], [0, 1], 'k--')
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel('False Positive Rate or [1 - True Negative Rate]')
plt.ylabel('True Positive Rate')
plt.title('Receiver operating characteristic example')
plt.legend(loc="lower right")
plt.show()
def draw_prec_recall(actual_values, probability_estimates):
p, r, thresholds = precision_recall_curve(actual_values, probability_estimates)
plt.plot(thresholds, p[:-1], "b-", label="Precision")
plt.plot(thresholds, r[:-1], "r-", label="Recall")
plt.title("Precison - Recall Trade off")
plt.legend(loc="lower right")
plt.show()
def get_metrics(y, pred, prob_est):
# Confusion Matrix
tn, fp, fn, tp = confusion_matrix(y, pred).ravel()
precision = precision_score(y, pred)
recall = recall_score(y, pred)
f = f1_score(y, pred)
# Sensitivity, Specificity
print(f"Sensitivity (Recall): {recall}\nSpecificity: {tn/(tn+fp)}\nPrecision: {precision}\nF-Score: {f}")
# Reciever Operating Characteristic Curve
draw_roc(y, prob_est[:, 1])
# Precision Recall Curve
draw_prec_recall(y, prob_est[:, 1])
get_metrics(y_train, pred, prob_est)
Finding Optimal Cut-Off¶
# Making y_train_pred_final
y_train_pred_final = pd.DataFrame({'Converted':y_train, 'probability': prob_est[:, 1]})
numbers = np.round(np.linspace(0,1,40,endpoint=False), 2)
for i in numbers:
y_train_pred_final[i]= y_train_pred_final.probability.map(lambda x: 1 if x > i else 0)
# Making cutoffs
cutoff_df = pd.DataFrame( columns = ['prob','accuracy','sensi','speci'])
num = np.round(np.linspace(0,1,40,endpoint=False), 2)
for i in num:
cm1 = confusion_matrix(y_train_pred_final.Converted, y_train_pred_final[i])
total1=sum(sum(cm1))
accuracy = (cm1[0,0]+cm1[1,1])/total1
speci = cm1[0,0]/(cm1[0,0]+cm1[0,1])
sensi = cm1[1,1]/(cm1[1,0]+cm1[1,1])
cutoff_df.loc[i] =[ i ,accuracy,sensi,speci]
cutoff_df['var'] = np.var([cutoff_df.accuracy, cutoff_df.sensi, cutoff_df.speci], axis=0)
# plotting accuracy sensitivity and specificity for various probabilities.
cutoff_df.plot.line(x='prob', y=['accuracy','sensi','speci'])
plt.show()
cutoff_value = cutoff_df.sort_values('var').head(1)['prob'].values[0]
print(f"Optimum cut-off value is: {cutoff_value}")
# new predicted values based on cut-off
pred = (log_reg.predict_proba(scaled_X_train[cols])[:, 1] >= cutoff_value).astype(int)
print(f"Accuracy: {accuracy_score(y_train, pred)}")
get_metrics(y_train, pred, prob_est)
Measuring Performance on Test Set¶
# Scoring against Test Set
log_reg.fit(scaled_X_train[cols], y_train)
pred = (log_reg.predict_proba(scaled_X_test[cols])[:, 1] >= cutoff_value).astype(int)
prob_est = log_reg.predict_proba(scaled_X_test[cols])
print(f"accuracy: {accuracy_score(y_test, pred)}")
get_metrics(y_test, pred, prob_est)
Finally, we have an overall accuracy of about 0.85 on our Logistic Regression model. That is, there is 85% chance that our predicted leads will be converted. This meets the CEO's target of atleast 80% lead conversion.
Lead Scoring¶
We will perform lead-scoring on the test set.
scores = pd.DataFrame({'lead_score':(prob_est[:, 1]*100).astype('int'), 'predicted_outcome': pred, 'actual_outcome': y_test}).reset_index(drop=True)
scores.head()